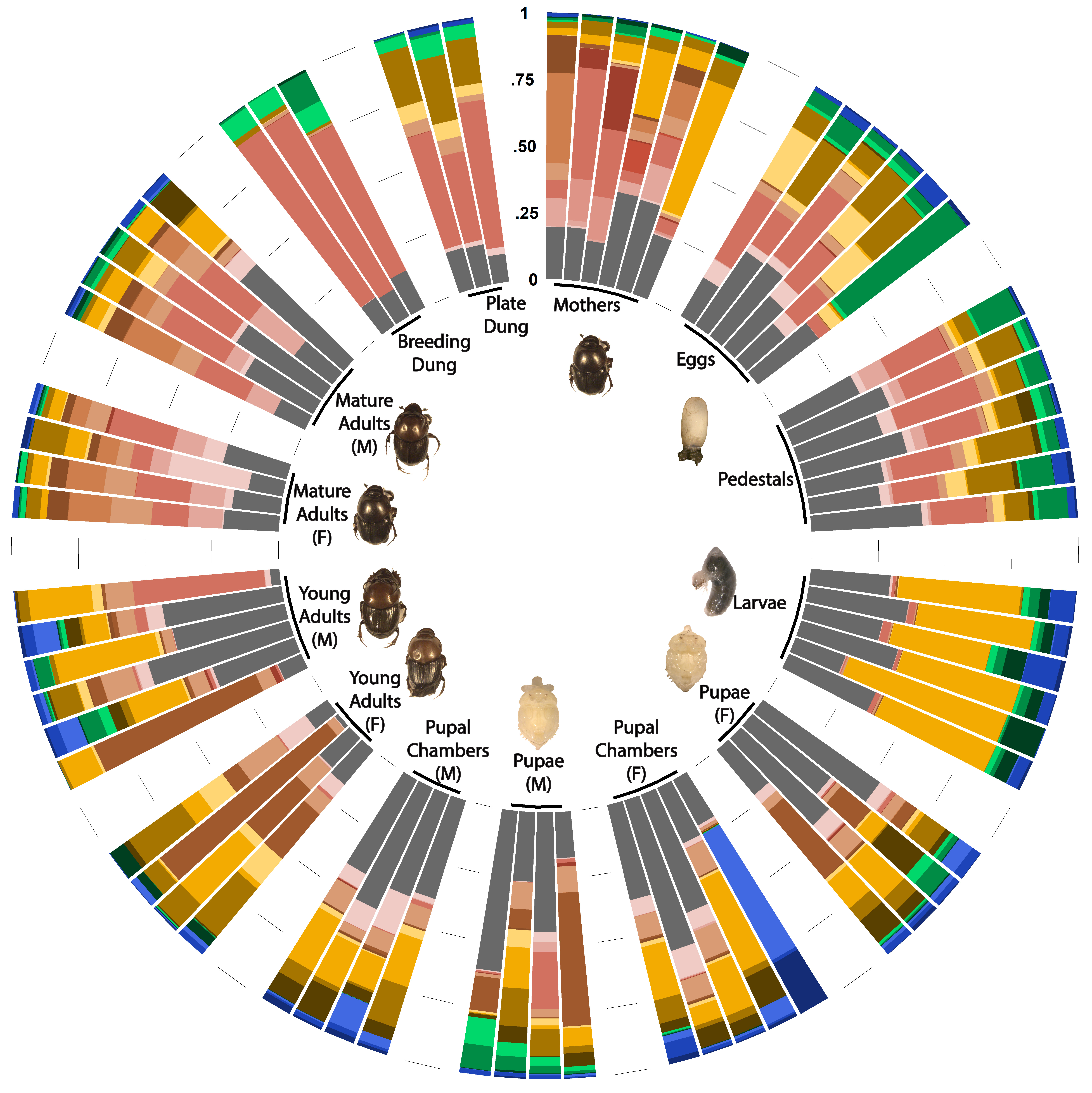
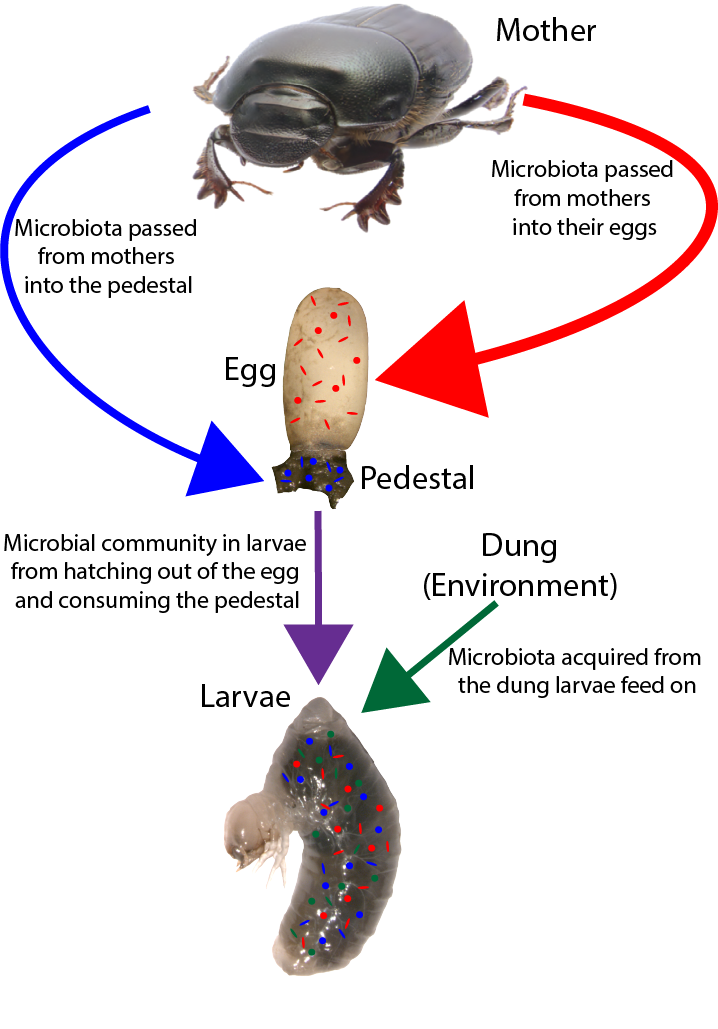
Influence of inherited microbiota on immunity
How may the inheritance of beneficial, or commensal, symbionts influence a host's ability to defend against and overcome infectious pathogens? How might environmental conditions influence any of these affects? Has host reliance on microbiome-mediated immune responses diversified across micro and/or macroevolutionary scales?
One specific benefit microbial partners may provide for their host is immune system stimulation. For example, studies suggest that supplementation of heritable microbiota to infants can significantly reduce conditions linked to immune system hypersensitivity, such as asthma and eczema (O’Neill et al., 2017), while increasing defense against potentially pathogenic invaders (Duar et al., 2020). Yet the mechanisms by which microbial associations interact with immune system components remain poorly understood outside few model systems and laboratory settings, and their contributions to host evolution and adaptation remain largely unexplored. I am currently working on a project to characterize the role of microbiota in determining the potential and functional immunity of an insect host and assessing the nature and degree of divergence among host species and populations in their reliance on microbiome-mediated immunity.